In 2024, the Nobel Prize in Chemistry recognized a transformative milestone in structural biology: the advent of AlphaFold2, a cutting-edge artificial intelligence (AI) system capable of predicting protein structures with unprecedented accuracy. This achievement, alongside rapid advances in protein language models, is reshaping our understanding of life at the molecular level. From unraveling fundamental biology to accelerating drug discovery, here’s a synthesized view of the revolution already underway and the exciting frontiers that lie ahead.
Why Protein Structures Matter
Proteins are the essential workhorses in all living cells, responsible for catalyzing reactions, transporting molecules, and orchestrating signaling pathways. Their unique 3D configurations—built from folded chains of amino acids—dictate their specific functions. For decades, scientists relied on labor-intensive experimental methods (like X-ray crystallography, NMR spectroscopy, or cryo-electron microscopy) to map these folds. While extremely precise, such techniques are time-consuming and expensive, leaving a vast gap between the millions of known protein sequences and the relatively small fraction of experimentally solved structures.
A 50-Year Challenge: Cracking the Protein Folding Code
Historically, the “protein folding problem” has been one of biology’s greatest puzzles. Researchers attempted to computationally determine a protein’s 3D shape solely from its amino acid sequence, an approach that required either:
Template-Based Modeling (TBM): Reliant on existing protein structures as templates.
Free Modeling (Ab Initio): Solely grounded in fundamental physics, generating myriad conformations.
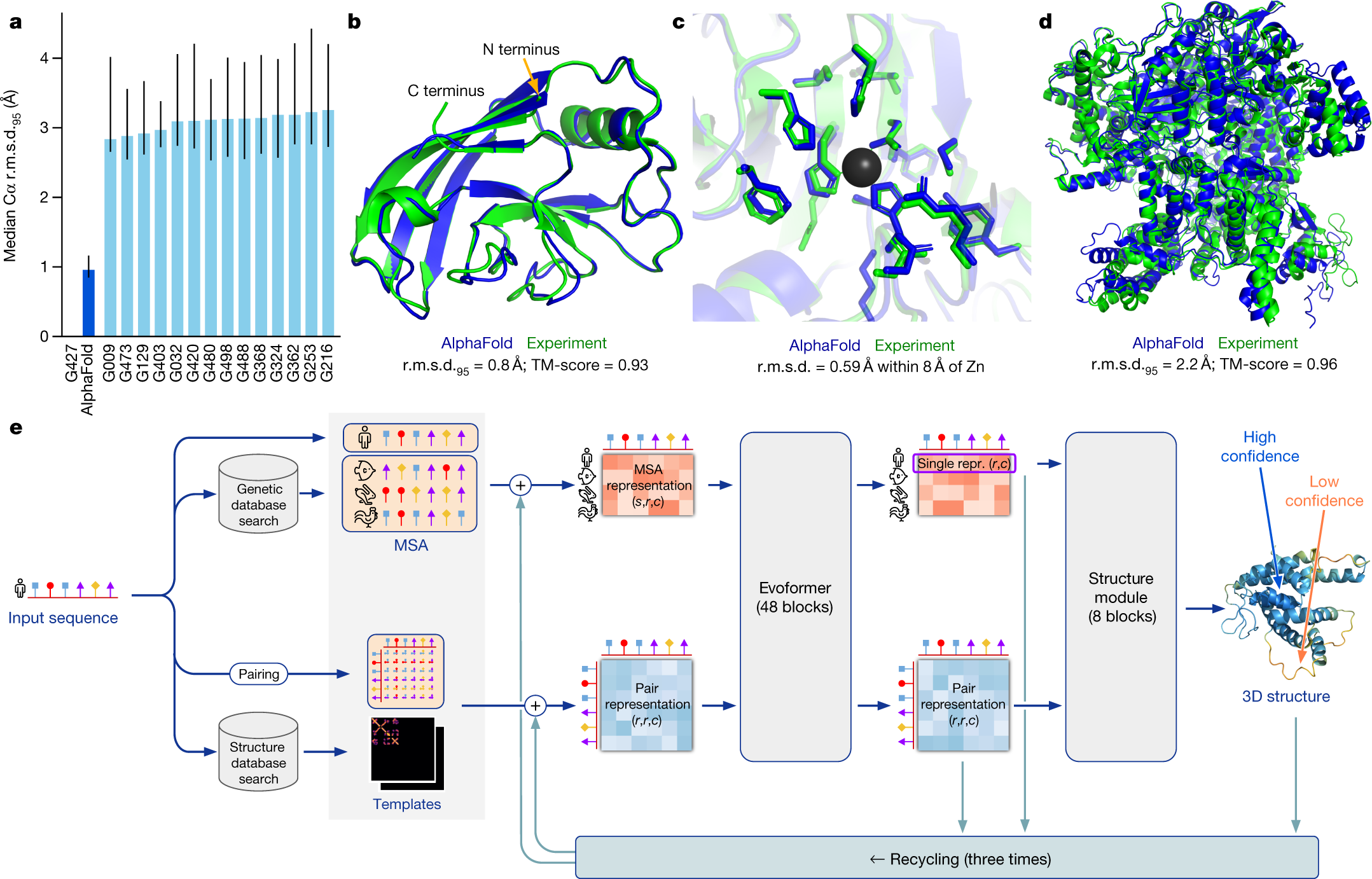
While these methods helped, they often fell short, especially for novel protein folds lacking close structural relatives. Enter AlphaFold2—a system that effectively overcame many of these hurdles and quickly garnered worldwide attention.
AlphaFold2: A Quantum Leap
Released by DeepMind in 2020 and later recognized by the Nobel Committee in 2024, AlphaFold2 revitalized the field with several core innovations:
Evoformer Architecture
Processes multiple sequence alignments (MSAs) to identify co-evolving amino acids.
Builds sophisticated “residue pair” representations, allowing the network to propose credible 3D structural hypotheses.
Structure Module
Employs equivariant transformers, ensuring that geometry is both precise and consistent regardless of how proteins are oriented in space.
Uses self-distillation: training on both experimental data and its own predictions to progressively refine accuracy.
CASP14 Triumph
Scored a median backbone accuracy around 0.96 Å—a near-atomic resolution—in the 2020 Critical Assessment of protein Structure Prediction competition.
Vastly outperformed other contemporary methods and set a new bar for what’s achievable with AI.
Within a short time, AlphaFold2’s predictions populated an online database of over 200 million protein structures, including a substantial portion of the human proteome.
Impact on Science and Medicine
Drug Discovery
Accelerated Screening: High-quality protein models allow rapid virtual screening, compressing what once took months into mere weeks. Notably, researchers identified a CDK20 inhibitor in under 30 days using AlphaFold-derived insights.
Personalized Medicine: Detailed maps of protein variants guide tailored therapies for genetic disorders or help predict drug resistance.
Bioengineering
Enzyme Design: By visualizing and tweaking catalytic sites, scientists can create enzymes that degrade plastic waste or produce biofuels more efficiently.
Novel Proteins: AI-driven design tools, such as ESMFold or EMBER3D, go beyond just predictions—enabling researchers to create entirely new proteins with specialized functions.
Structural Biology
Complexes and Large Assemblies: Hybrid approaches (e.g., AlphaFold + cryo-EM) clarify enormous molecular machines—like ribosomes or viral spike proteins.
Disease Mechanisms: Accurate models of proteins implicated in conditions like Alzheimer’s or COVID-19 can spotlight mutation effects and inform strategies for vaccine or drug design.
Toward AlphaFold3 and Extended Possibilities
Building on AlphaFold2, AlphaFold3 employs diffusion-based methods to improve ligand binding accuracy and handle more complex scenarios:
Protein–Nucleic Acid Complexes: Predicts how proteins interact with DNA or RNA, crucial for understanding transcription regulation or virus replication.
Small Molecule Docking: Advances the design of therapeutics by fine-tuning our view of how drugs bind (or fail to bind) a protein target.
Modified Residues & Ligands: Accommodates non-standard amino acids and covalent bonds, mirroring the real-world chemistry often overlooked in earlier AI models.
This progression underscores a broader trend: AI is not just refining single-protein predictions but mapping multi-chain complexes, post-translational modifications, and ever more dynamic conformations.
Limitations and Future Directions
Despite the enormous strides, challenges remain:
Disordered Regions & Dynamics
AI predictions typically yield static snapshots, whereas many proteins naturally switch shapes to function.
Flexible loops or intrinsically disordered segments require advanced modeling approaches that track shape changes over time.
Massive Assemblies & Real-World Complexity
Systems such as viruses or eukaryotic super-complexes push computational tools to their limits.
Realistic modeling involves not just proteins but also environmental factors, membranes, and varying pH conditions.
Ethical and Open Science Considerations
Proprietary or opaque models may limit global research access.
Ensuring equitable distribution of these cutting-edge tools fosters innovation across disciplines and geographies.
Still, each new release—backed by open-source data and robust research collaborations—brings the scientific community closer to handling these puzzles.
Conclusion: A Paradigm Shift in Protein Science
From its first near-perfect predictions at CASP14 to its Nobel recognition in 2024, AlphaFold2 has proven that AI can crack some of biology’s toughest puzzles. We now live in a digital biology era where computational models and experimental data interlace, powering breakthroughs at speeds once deemed impossible.
Looking Ahead, expect deeper integration with lab-based experiments, expansions into predicting dynamic protein states, and widespread adoption of AI in fields like agriculture, climate science, and synthetic biology. By combining the creativity of machine learning with the rigor of experimental validation, we stand on the cusp of uncovering life’s molecular secrets at an unprecedented scale.
“It’s not just a tool; it’s a paradigm shift,” as many leading researchers echo. From my readings, the excitement is palpable: AI-driven models have already changed the questions we ask in biology, pointing us toward a horizon where the unknown quickly becomes the newly discovered.
Where to Go Next?
Explore Databases: Dive into the AlphaFold Database to find structures relevant to your research questions.
Collaborate: Partner with labs using cryo-EM or NMR to refine and confirm AI models for complex protein assemblies.
Experiment & Build: If you’re in synthetic biology, harness these predictive tools to engineer proteins or enzymes tailored to your needs—whether for drug development or environmental solutions.
Share Insights: Contribute back to open repositories, ensuring continued innovation and global access to these groundbreaking methods.
Ultimately, AlphaFold2 and beyond is more than just a milestone in structural prediction—it's a leap toward understanding and shaping the very foundations of life itself.